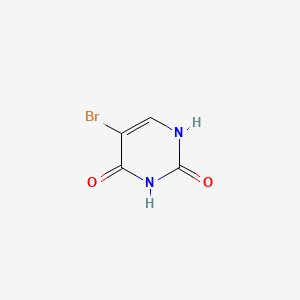

1. 5 Bromouracil
2. Bromouracil
3. Bromuracil
1. 51-20-7
2. Bromouracil
3. 5-bromopyrimidine-2,4(1h,3h)-dione
4. 5-bromo-1h-pyrimidine-2,4-dione
5. 5-bromo-2,4(1h,3h)-pyrimidinedione
6. Uracil, 5-bromo-
7. 5-bromo-2,4-dihydroxypyrimidine
8. 2,4(1h,3h)-pyrimidinedione, 5-bromo-
9. 5-bromopyrimidine-2,4-diol
10. Mfcd00006017
11. 5-bromo-1,2,3,4-tetrahydropyrimidine-2,4-dione
12. Chebi:20552
13. 4hk400g5uo
14. 1,2,3,4-tetrahydro-5-bromo-2,4-pyrimidinedione
15. Nsc-19940
16. 5-bru
17. Einecs 200-084-0
18. Nsc 19940
19. Unii-4hk400g5uo
20. Uracil, 5-bromo- (van)
21. 5-bu
22. Ai3-25471
23. 5-bromo-uracil
24. Hsdb 7495
25. Bromhydroxypyrimidinon
26. 5-bromouracil, 98%
27. Epitope Id:138109
28. 5-brom-6-hydroxypyrimidinon
29. 5-bromouracil [mi]
30. Schembl29805
31. Chembl144730
32. Schembl8740942
33. Dtxsid2058758
34. Zinc967318
35. 2,3h)-pyrimidinedione, 5-bromo-
36. Act04613
37. Nsc19940
38. Str01501
39. Bbl013052
40. Ccg-46723
41. Stk301806
42. Stl451516
43. Akos001448866
44. Akos009097562
45. Ab00570
46. Ac-3034
47. Am83917
48. Cs-w001982
49. Bp-12090
50. Sy003631
51. 5-bromo-2,4(1h,3h)-pyrimidinedione #
52. Db-002591
53. B0665
54. Eu-0035288
55. Ft-0602493
56. En300-51230
57. B-9200
58. 006b017
59. Ac-907/25014019
60. Q238477
61. 2,4-pyrimidinedione, 1,2,3,4-tetrahydro-5-bromo
62. Sr-01000636400-1
63. F3096-1702
64. Z275128628
65. Urb
| Molecular Weight | 190.98 g/mol |
|---|---|
| Molecular Formula | C4H3BrN2O2 |
| XLogP3 | -0.2 |
| Hydrogen Bond Donor Count | 2 |
| Hydrogen Bond Acceptor Count | 2 |
| Rotatable Bond Count | 0 |
| Exact Mass | 189.93779 g/mol |
| Monoisotopic Mass | 189.93779 g/mol |
| Topological Polar Surface Area | 58.2 Ų |
| Heavy Atom Count | 9 |
| Formal Charge | 0 |
| Complexity | 199 |
| Isotope Atom Count | 0 |
| Defined Atom Stereocenter Count | 0 |
| Undefined Atom Stereocenter Count | 0 |
| Defined Bond Stereocenter Count | 0 |
| Undefined Bond Stereocenter Count | 0 |
| Covalently Bonded Unit Count | 1 |
Antimetabolite
National Library of Medicine's Medical Subject Headings online file (MeSH, 2007)
/EXPTL THER/ The ternary complexes of Mn(II), Co(II), Ni(II), Cu(II), Zn(II), and Cd(II) ions with 5-halouracils, viz., 5-fluorouracil (5FU), 5-chlorouracil (5ClU), and 5-bromouracil (5BrU), and the biologically important ligand L-histidine (HISD) have been synthesized and characterized by elemental analysis, conductance measurements, infrared spectra, electronic spectra, and magnetic moment (room temperature) measurements. On the basis of these studies, the structures of the complexes have been proposed. All these ternary complexes were screened for their antitumor activity against Dalton's lymphoma in C3H/He mice. It was found that only Mn(II)-5BrU-HISD, Co(II)-5BrU-HISD, Cu(II)-5ClU-HISD, Cu(II)-5BrU-HISD, Zn(II)-5FU-HISD, and Zn(II)-5BrU-HISD complexes have significant antitumor activity with T/C greater than 125% (where T and C represent mean lifespan of treated mice and control mice respectively). The Mn(II)-5FU-HISD, Co(II)-5FU-HISD, Co(II)-5ClU-HISD, Ni(II)-5ClU-HISD, Ni(II)-5BrU-HISD, and Zn(II)-5ClU-HISD complexes are also effective antitumor agents, with T/C greater than 115%. The complexes that showed effective antitumor action in vivo were also found to inhibit 3H-thymidine incorporation (DNA replication) in Dalton's lymphoma cells in vitro.
PMID:2628549 Singh UP et al; J Inorg Biochem 37 (4): 325-39 (1989)
Antimetabolites
Drugs that are chemically similar to naturally occurring metabolites, but differ enough to interfere with normal metabolic pathways. (From AMA Drug Evaluations Annual, 1994, p2033) (See all compounds classified as Antimetabolites.)
Mutagens
Chemical agents that increase the rate of genetic mutation by interfering with the function of nucleic acids. A clastogen is a specific mutagen that causes breaks in chromosomes. (See all compounds classified as Mutagens.)
Eosinophils use eosinophil peroxidase, hydrogen peroxide (H(2)O(2)), and bromide ion (Br(-)) to generate hypobromous acid (HOBr), a brominating intermediate. This potent oxidant may play a role in host defenses against invading parasites and eosinophil-mediated tissue damage. In this study, /the authors/ explore the possibility that HOBr generated by eosinophil peroxidase might oxidize nucleic acids. When /the authors/ exposed uracil, uridine, or deoxyuridine to reagent HOBr, each reaction mixture yielded a single major oxidation product that comigrated on reversed-phase HPLC with the corresponding authentic brominated pyrimidine. The eosinophil peroxidase-H(2)O(2)-Br(-) system also converted uracil into a single major oxidation product, and the yield was near-quantitative. Mass spectrometry, HPLC, UV--visible spectroscopy, and NMR spectroscopy identified the product as 5-bromouracil. Eosinophil peroxidase required H(2)O(2) and Br(-) to produce 5-bromouracil, implicating HOBr as an intermediate in the reaction. Primary and secondary bromamines also brominated uracil, suggesting that long-lived bromamines also might be physiologically relevant brominating intermediates. Human eosinophils used the eosinophil peroxidase-H(2)O(2)-Br(-) system to oxidize uracil. The product was identified as 5-bromouracil by mass spectrometry, HPLC, and UV--visible spectroscopy. Collectively, these results indicate that HOBr generated by eosinophil peroxidase oxidizes uracil to 5-bromouracil. Thymidine phosphorylase, a pyrimidine salvage enzyme, transforms 5-bromouracil to 5-bromodeoxyridine, a mutagenic analogue of thymidine. These findings raise the possibility that halogenated nucleobases generated by eosinophil peroxidase exert cytotoxic and mutagenic effects at eosinophil-rich sites of inflammation.
PMID:11329272 Henderson JP et al; Biochemistry 40 (7): 2052-9 (2001)
... Using a sensitive and specific mass spectrometric method, /the authors/ detected two products of myeloperoxidase, 5-chlorouracil and 5-bromouracil, in neutrophil-rich human inflammatory tissue. Myeloperoxidase is the most likely source of 5-chlorouracil in vivo because halogenated uracil is a specific product of the myeloperoxidase system in vitro. In contrast, previous studies have demonstrated that 5-bromouracil could be generated by either eosinophil peroxidase or myeloperoxidase, which preferentially brominates uracil at plasma concentrations of halide and under moderately acidic conditions. These observations indicate that the myeloperoxidase system promotes nucleobase halogenation in vivo. Because 5-chlorouracil and 5-bromouracil can be incorporated into nuclear DNA, and these thymine analogs are well known mutagens, our observations raise the possibility that halogenation reactions initiated by phagocytes provide one pathway for mutagenesis and cytotoxicity at sites of inflammation.
PMID:12707270 Henderson JP et al; J Biol Chem 278 (26): 23522-8 (2003)
/Among/ halogenated pyrimidines ... if one compares the van der Waals radii of the various 5-position substituents, the dimension of the fluorine atom resembles that of hydrogen /ie, uracil/, whereas the bromine and iodine atoms are larger and close in size to the methyl group /ie, thymine/ ... /Pyrimidine analogs/
Hardman, J.G., L.E. Limbird, P.B., A.G. Gilman. Goodman and Gilman's The Pharmacological Basis of Therapeutics. 10th ed. New York, NY: McGraw-Hill, 2001., p. 1404
Cultivation of E. coli cells in the presence of 5-bromodeoxyuridine (BUdR) leads to formation of lesions in the cellular DNA which affect its secondary structure, as reflected by changes in temperature profiles. Such DNA contains single-stranded regions susceptible to endonuclease S1. One of the major sources of the BU-induced lesions appears to be dehalogenation of incorporated 5-bromouracil (BU) residues, with accompanying formation of uracil. The presence of uracil residues in such DNA was demonstrated directly by chromatography of hydrolyzates, and by the susceptibility of such residues to uracil-DNA glycosylase. The number of uracil residues was dependent on the extent of damage in the DNA, and decreased during the DNA repair that accompanied reactivation of bromouracil-inactivated cells. Dehalogenation of incorporated BU presumably results in formation of apyrimidinic sites by uracil-DNA glycosylase, and then single-strand nicks either by AP-endonuclease and/or dehalogenation. The findings are relevant to the mechanism of BU-induced mutagenesis.
PMID:6188038 Szysko J et al; Mutat Res 108 (1-3): 13-27 (1983) .
The early studies are recounted, that led to the discovery of the ubiquitous process of DNA excision repair, followed by a review of the pathways of transcription-coupled repair (TCR) and global genomic nucleotide excision repair (GGR). Repair replication of damaged DNA in UV-irradiated bacteria was discovered through the use of 5-bromouracil to density-label newly synthesized DNA. This assay was then used in human cells to validate the phenomenon of unscheduled DNA synthesis as a measure of excision repair and to elucidate the first example of a DNA repair disorder, xeroderma pigmentosum. Features of the TCR pathway (that is defective in Cockayne syndrome (CS)) include the possibility of "gratuitous TCR" at transcription pause sites in undamaged DNA. The GGR pathway is shown to be controlled through the SOS stress response in E. coli and through the activated product of the p53 tumor suppressor gene in human cells. These regulatory systems particularly affect the efficiency of repair of the predominant UV-induced photoproduct, the cyclobutane pyrimidine dimer, as well as that of chemical carcinogen adducts, such as benzo(a)pyrene diol-epoxide. Rodent cells (typically lacking the p53-controlled GGR pathway) and tumor virus infected human cells (in which p53 function is abrogated) are unable to carry out efficient GGR of some lesions. Therefore, caution should be exercised in the interpretation of results from such systems for risk assessment in genetic toxicology. ...
PMID:11341989 Hanawalt PC; Mutat Res 485 (1): 3-13 (2001)
The incorporation of bromouracil into the deoxyribonucleic acid (DNA) of bacteria and viruses has been explained on the basis that bromouracil is similar to thymine in its ability to form hydrogen bonds with adenine. Enzymatic experiments support this interpretation. Bromouracil incorporation into DNA proceeds via bromouracil deoxynucleoside triphosphate, which is an effective substitute for thymine deoxynucleoside triphosphate in the replication of DNA. Results with this and other base analogues have shown that the enzymatic replication of DNA is governed by pairing of a 6-aminopyrimidine to a 6-ketopurine (e.g., cytosine to guanine) and of a 6-ketopyrimidine to a 6-aminopurine (e.g., thymine to adenine). Several questions might be raised about this generalization. First, the accuracy previously achieved in measuring in vitro incorporation disclosed deviations from this rule only when they occurred with a frequency greater than 0.02 per cent. Would a more sensitive technique reveal pairing errors occurring at an even lower frequency? Second, the incorporation of bromouracil into viral or bacterial DNA is associated with an increase in the mutation rate; mutations due to occasional pairing of guanine with tautomeric forms of thymine or bromouracil that resemble cytosine has been suggested by Watson and Crick and Freese. Matching of bromouracil with guanine instead of with adenine leads, in subsequent replications, to a G-C pair in place of an original A-T. Would the presence of bromouracil in a DNA primer increase the incidence of "incorporation errors" when the DNA is enzymatically replicated? Finally, Shapiro and Chargaff determined the yield of pyrimidine nucleotides and oligonucleotides after acid hydrolysis of bromouracil-substituted Escherichia coli DNA and concluded that the inclusion of bromouracil in DNA led to drastic changes in the over-all arrangement of the bases. It was therefore important to check this conclusion by another method, for example, by analysis of nearest-neighbor base sequences. The availability of a DNA-like polymer made up exclusively of A and T arranged in alternating sequence (dAT polymer)7 made possible the synthesis of the analogous dABU polymer. These polymers are ideal primers for studying the mismatched incorporation of G residues. Using an assay that would have revealed one G residue per 105 A and T nucleotides polymerized, /the authors/ failed to observe G incorporation in dAT-primed reactions. However, with dABU as primer, the incorporation of G was unequivocal; it occurred at frequencies ranging from 1 per 2,000 to 1 per 25,000 nucleotides polymerized. These "errors" induced by BU are 2 to 3 orders of magnitude too low in frequency to account for the nucleotide disarrangement reported by Shapiro and Chargaff. /The authors/ therefore determined the nucleotide sequences in bromouracil-substituted E. coli DNA by the nearest-neighbor analysis; arrangement of nucleotides was indistinguishable from that in the normal sample. Enzymatic replication of a DNA-like polymer which contains only adenine and thymine residues (dAT polymer) revealed no detectable incorporation of guanine residues. The level of sensitivity of these experiments shows the frequency of "aberrant" guanine incorporation to be less than one residue per 28,000-580,000 adenine and thymine nucleotides polymerized. In the replication of an analogous polymer containing bromouracil in place of thymine (dABU polymer), the incorporation of guanine was unequivocal. It occurred at a frequency of one per 2,000 to 25,000 adenine and thymine nucleotides polymerized. Analysis of the sequential arrangement of the incorporated guanine residues in the synthesized product showed the nearest neighboring base to be bromouracil, guanine, and adenine with frequencies of 41, 42, and 17%, respectively. Current theories /in 1962/ of bromouracil mutagenesis, if applied to the replication of this polymer, would have predicted the incorporation of guanine exclusively next to bromouracil. The nearest-neighbor nucleotide sequences of normal E. coli DNA were compared with those of bromouracil-substituted E. coli DNA and found to be indistinguishable (-42%). This analysis does not support a contention based on results from acid hydrolysis of DNA that drastic changes in sequence result from substitution of bromouracil in E. coli DNA.
PMID:13922323 Full text: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC220799 Trautner TA et al; PNAS 48:;449-55 (1962)
... The radiosensitization properties of BrdUrd result primarily from the electrophilic nature of the bromine, making it a good leaving group and leading to the irreversible formation of the uridine-yl radical (dUrd(.)) or the uridine-yl anion (dUrd(-)) upon addition of an electron. The radiolytic loss of the bromine atom is greatly suppressed in double-stranded compared to single-stranded DNA. Thus /the authors/ propose that the radiosensitization effects of bromouracil in vivo will likely be limited to single-strand regions such as found in transcription bubbles, replication forks, DNA bulges and the loop region of telomeres. /The authors believe that their /results may have profound implications for the clinical use of bromodeoxyuridine (BrdUrd) as a radiosensitizer as well as for the development of targeted radiosensitizers.
PMID:15548110 Cecchini S et al; Radiat Res 162 (6): 604-15 (2005)